MWLC Phone: 317-236-2099
https://madamwalkerlegacycenter.com/
This event provided a wonderful opportunity to connect with peers, engage with faculty, and enjoy the vibrant LSAMP community.
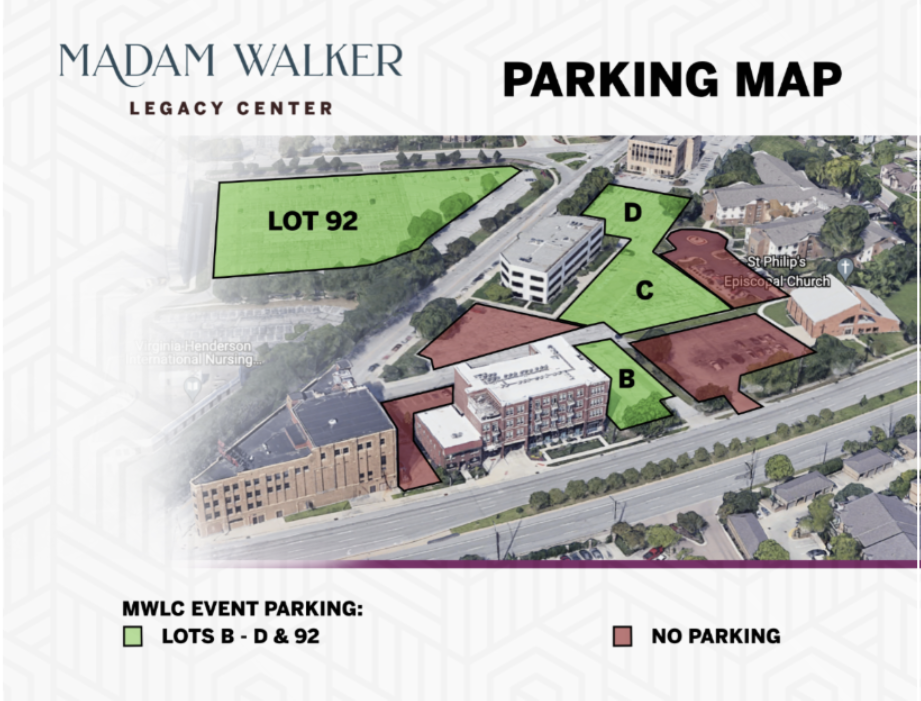

MWLC Phone: 317-236-2099
https://madamwalkerlegacycenter.com/
This event provided a wonderful opportunity to connect with peers, engage with faculty, and enjoy the vibrant LSAMP community.

11:00-11:15 a.m.: Arrival, registration, and poster set-up (for session # 1)
11:15 a.m.-Noon: Poster Session #1
Upon arrival, poster presenter(s) will be directed to their poster board.
*Please note: Information about student poster presentations, including title and description, was provided by the individual student and/or their mentor(s).
Human Serum Albumin Nanoparticles for Enzymatic Delivery of Rhein and Infrared Ionic Drug Against Cancer
Name: Garcia-Garcia, Diana
Email: diana.garciagarcia@bsu.edu
Campus: BSU
Faculty Advisor: Dr. David Bwambok
Faculty Advisor's Email: david.bwambok@bsu.edu
Chemotherapy has been the main cancer treatment focus. It can limit tumor progression and can target non-cancerous cells. The issue with chemotherapy is that it is not very selective. This research focuses on the viability of Rhein through a multifunctional system, a chemotherapeutic agent that can allow selectivity through nanoparticles as well as testing the compatibility to IR780BETI dye for NIRF imaging in cancer and photothermal therapy through nanoparticles.
Synthesis of Human Serum Albumin Nanoparticles for Enzymatic Delivery of Camptothecin and Infrared Ionic Drug Against Cancer
Name: Galindo, Erica
Email: ehgalindo@bsu.edu
Campus: BSU
Faculty Advisor: Dr. David Bwambok
Faculty Advisor's Email: david.bwambok@bsu.edu
Cancer is a leading cause of death globally. Early diagnosis and treatment are crucial for improving therapeutic outcomes. Traditional treatments have limitations, including incomplete surgical resection and toxicity on noncancerous cells. To address these limitations, this study aims to develop a hybrid drug delivery system that consists of a chemotherapeutic agent, Camptothecin (CPT), and a photosensitizing agent, IR780BETI. Human serum albumin (HSA) nanoparticles (NPs) were used for drug delivery to leverage the enhanced permeability and retention (EPR) effect for targeted tumor therapy. The hybrid CPT-HSA-IR780BETI-NPs were synthesized by desolvation and crosslinked with 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) for drug encapsulation. The size of the NPs was 141.5 nm, as determined by dynamic light scattering (DLS), indicating an appropriate size for EPR effect-induced tumor targeting. The zeta potential of these NPs was -19.12 mV, suggesting relative stability to aggregation. The drug release from the HSA-NPs was assessed using absorbance and fluorescence measurements at various time intervals after treatment with collagenase, simulating the enzymatic release of the target drugs in an in vitro tumor environment. This system offers promising advancements in cancer treatment by providing targeted delivery, simultaneous diagnosis, and three cancer therapies.
Investigation of Enantiomeric Recognition for Pharmaceutical Quality Control using Dipeptide Fluorescent Chiral Ionic Liquids
Name: Hughes, Shianna
Email: shianna.hughes@bsu.edu
Campus: BSU
Faculty Advisor: Dr. Bwambok
Faculty Advisor's Email: david.bwambok@bsu.edu
About 50% of drugs are chiral, while the other 50% are racemic. Enantiomers of chiral drugs can vary in safety, effectiveness, and pharmaceutical properties. For instance, the R-enantiomer of thalidomide treated morning sickness, but the S-enantiomer caused birth defects. Therefore, enantiomer screening is crucial before drugs reach the market. This research aims to design a method using dipeptide-based chiral ionic liquids to screen and quantify drug purity. The method employs fluorescence, UV spectroscopy, and NMR to differentiate enantiomers. Prolylphenylalanine (PPA) dipeptide-based chiral ionic liquid is used for its dual chiral centers and fluorescence properties, ensuring medications meet purity and potency standards.
Optimized Synthesis of Cinnamic Acid Derivatives
Name: Johnson, Dre'An
Email: drean.johnson@bsu.edu
Campus: BSU
Faculty Advisor: Robert Sammelson
Faculty Advisor's Email: resammelson@bsu.edu
Researching and optimizing different methods and procedures of synthesizing a variety of cinnamic acid derivatives. These cinnamic acid derivatives are a small part of a larger ongoing research project in which they are connected to a larger compound called Ipomoeassin F. Ipomoeassin F is being studied in breast cancer medication research due to its anticancer properties, most of which being affected by the cinnamate group present in the compound.
Investigation of the Mechanisms of Coronary Vessel Development: Genotyping of the Transgenic Mouse used in the Study
Name: Kusasira, Esther
Email: esther.kusasira@bsu.edu
Campus: BSU
Faculty Advisor: Dr. Bikram Sharma
Faculty Advisor's Email: bsharma@bsu.edu
Dr. Sharmas lab focuses on studying the mechanisms behind coronary vessels. Study the mechanisms of coronary vessels will aid in helping invent better treatment options for patients with cardiovascular diseases. My part in the lab was to genotype the mice that were being used to study the coronary vessels.
Updating Legacy Code
Name: Pena, Santos
Email: santos.pena@bsu.edu
Campus: BSU
Faculty Advisor: David Largent
Faculty Advisor's Email: dllargent@bsu.edu
I am working on updating a mediaComp library so that it can work on different IDEs. An IDE is a software where new programmers use to learn to code. It has functions and tips that could help a new programmer get started because it can be very overwhelming when starting out. The mediaComp library is the code inside an IDE and makes all the functions and tips possible. I was tasked with taking The mediaComp from an IDE called JES and then trying to transfer its functions into a new IDE. My first steps were to learn how the media comp works by using JES as a learning tool. Then after that I would have to learn how I could possibly transfer the code to a new IDE in a simple process that anyone could do. This would also require me to fix some errors in the code because it is written in Python 2 and we need to get it to python 3.
Identification of Nuclear Localization Sequences in C. Albicans Pseudouridine Synthase 1
Name: Sanders, Mercedes
Email: mercedes.sanders@bsu.edu
Campus: BSU
Faculty Advisor: Douglas Bernstein
Faculty Advisor's Email: dabernstein@bsu.edu
Pseudouridine synthase 1, also known as Pus1, is an enzyme that converts uridine into pseudouridine after the nucleotide has been incorporated into RNA. Genome-wide expression studies of the diploid human fungal pathogen Candida albicans indicate that CaPUS1 is induced during biofilm formation, a key virulence factor. After translation, some Pus1 localizes to the nucleus, however, the Bernstein lab has found Pus1 distribution changes under stress. Clusters of positively charged amino acids have been shown to be crucial for nuclear import in other species. The aim of this project is to find amino acid sequences important for localization to the nucleus in C. albicans. CRISPR/Cas9-mediated genome editing will be applied to target a stretch of 8 consecutive positively charged amino acids in GFP-tagged Pus1. Visualization of the transformants to determine localization of Pus1 will be done via fluorescence microscopy. If the modified amino acid cluster is important for nuclear import, the fluorescence microscopy will show an increase of GFP in the cytoplasm.
Improving Particle Identification in Realistic Heavy Ion Collisions
Name: Washington, Trinity
Email: Trinity.Washington@bsu.edu
Campus: BSU
Faculty Advisor: Dr. Micheal Skoby
Faculty Advisor's Email: mjskoby@bsu.edu
In this project, I will use machine learning to understand how the strong force, which holds the nucleus together and also holds quarks and gluons together to make protons and neutrons by analyzing data provided by my mentor Dr. Micheal Skoby. I’ll be studying the state of matter called the quark-gluon plasma. When these quarks and gluons interact with each other through the strong force, they make a large number of new quarks and gluons. As the system cools, the quarks and gluons recombine to make various species of particles which are observed by particle detectors. Particle species are defined by their quark content, for example, a pion is made of an up quark and anti down quark. This helps us understand how the strong force makes new quarks, it is important to identify the species of particles that come from the collision. The goal of this project is to improve the quality of particle identification in these collisions using advanced machine learning techniques such as deep neural networks. We will use simulated particles and process them through a simulated particle detector to train and test the machine learning algorithm and compare its performance to traditional techniques of particle identification.
Investigation of the Radioactive Decay of Radon Using Python Software
Name: Williams, Tierra
Email: tierra.williams@bsu.edu
Campus: BSU
Faculty Advisor: Jennifer Coy
Faculty Advisor's Email: jennifer.coy@bsu.edu
In this project, I will be using the data analysis capabilities of Python to perform further research and analysis on how and why the element radon is decaying in an unexpected pattern. Radon is a naturally occurring element that regularly breaks down into other elements. But, when we breathe it in, it breaks down into elements that are solids, so those get trapped in our lungs. Because of this, a Geological Survey of Israel (GSI) was set up to study the decay of radon, which in theory should happen on a consistent basis according to the half-life of the element. However, it was found to decay on a more inconsistent basis than was previously thought. In my research, I am using Python to analyze and provide graphical representations of the GSI data through the years of 2008 to 2017. Of the seven columns of data collected, only one has been truly studied. So, my research focuses on graphing the other columns of data into 10 single year graphs per column, as well as one amalgamized graph with all of the years on one graph per column.
fNIR Systems Applied in Lie Detection
Name: Oduwole, Temi
Email: Ouatdale@gmail.com
Campus: IUB
Faculty Advisor: Melissa Blunck
Faculty Advisor's Email: Melhorto@iu.edu
Functional near-infrared spectroscopy (fNIRs) primarily measures the hemodynamic responses in the prefrontal cortex, a region associated with executive functions, including decision-making and inhibition. In the "To Tell the Truth" study, we investigate an individual's ability to detect a lie from the truth using a NIRs device and measuring blood oxygen levels associated with watching videos where people are both lying and telling the truth.
The Effects of Cyanogenesis on Flower Size
Name: Stevenson, Layla
Email: Laysteve@iu.edu
Campus: IUB
Faculty Advisor: Dr. Jennifer Lau
Faculty Advisor's Email: jenlau@iu.edu
Dr. Jen Lau and the graduate team helped me when deciding on my independent project. My project consisted of studying the effects of cyanogenesis on white clovers. I was looking into how does the cyanide being used a defense mechanism against herbivores affect their reproductive traits like their flower size. I devised a question and method to help analyze my results and think about future directions.
Revisiting HaptyBird: Integrating Unity and Logitech Racing Wheels for Improved User Interaction and Feedback
Name: Jorge, Ronaldo
Email: rjjorge@iu.edu
Campus: IUB
Faculty Advisor: Dr. Gregory Lewis
Faculty Advisor's Email: lewigr@iu.edu
In this project, we redesign the HaptyBird game by transitioning from Python to Unity
and incorporating Logitech G Pro racing wheels for enhanced haptic feedback. This upgrade
aims to improve the game’s graphical interface and user experience, facilitating intuitive control
as players steer a ball through obstacles. Preliminary tests indicate that the Unity-based
HaptyBird enhances user engagement, potentially yielding richer data on trust and cooperation
dynamics in gaming interactions.
A Study on GPR119 and Its Role In Weight Metabolism
Name: Garcia, Catalina
Email: garcikat@iu.edu
Campus: IUB
Faculty Advisor: Dr. Alex Straiker & Dr. Ken Mackie
Faculty Advisor's Email: straiker@iu.edu; kmackie@iu.edu
In 2013 about 7% of American adults were cannabis users and in 2021 that number increased to 12%, making it more common for adults to use cannabis. Frequent cannabis users often exhibit lower body weight, reduced BMIs, and a decreased likelihood of developing type 2 diabetes. Cannabis usage causes a side effect known as the “munchies” which causes a craving for food while intoxicated. If frequent cannabis users get the “munchies” and crave more food while intoxicated their weight should go up, right? That doesn’t seem to be the case though.GPR119 is a G-coupled protein receptor and a potential target for treating diabetes. Activation of this receptor has been shown to stimulate the release of insulin and glucagon-like peptide 1 (GLP-1) which act in lipid and glucose metabolism and the suppression of appetite. GPR119 is expressed in pancreatic β cells and intestinal enteroendocrine cells. It is a fat sensor activated by endogenous ligands such as OEA and Oleoyl-lysophosphatidylcholine and recently our lab has found that GPR119 can be activated by Δ9-tetrahydrocannabinol (THC), the psychoactive component of cannabis. RT-qPCR was used in this study on livers or intestines from obese GPR119 knockout (KO) mice that were treated with either vehicle or THC. This analytical method aimed to show changes in expression levels of genes involved with inflammation, lipogenesis, and glucose metabolism. Results from intestine show that all genes trend towards increased expression with THC treatment. GLP-1 and GLP-2 are involved in weight loss and glucose metabolism, whereas TNFα is involved in inflammation. Results from liver show that MGL increases with THC treatment, whereas DGLα and NAPE do not show an increased trend. MGL is an important enzyme that hydrolyzes the endocannabinoid, 2-AG. The changes observed in GPR119 KOs will be compared with results obtained from wild-type animals to better understand how THC may work through this receptor to modulate metabolism in the obese mouse.
Examination of Growth Rate In Rhizobia Strain Groups
Name: Virgen, Alexa
Email: aivirgen@iu.edu
Campus: IUB
Faculty Advisor: Dr. Jennifer Lau
Faculty Advisor's Email: aivirgen@iu.edu
Rhizobia are a group of bacteria that live on plant roots and form symbiotic relationships
with a variety of plant species in the legume family. The bacteria fix nitrogen into a usable form
for the plant, and the plant provides carbohydrates for bacteria. This relationship occurs in a
nodule on a plant root where the bacteria live. Legume-rhizobia interactions are affected by
nitrogen addition. Past work has shown rhizobia evolve to be worse mutualists in high nitrogen,
but other traits such as growth rate may have also changed. The aim of the experiment was to
determine how these rhizobia strains differ in growth rate. The genes controlling the rhizobium’s
interaction with the plant are housed on a plasmid. The rhizobia studied come from two separate
plasmid groups, as well as a few strains that have lost the plasmid. The rhizobia were grown
from frozen cultures, and incubated for 48 hours before quantifying growth. Growth was
measured using optical density measurements. Differences between plasmid groups and
differences between rhizobia that evolved in high nitrogen vs. low nitrogen environments were
analyzed using R. Future experiments could involve exposing strains to stress and seeing how
different strains respond, and how that relates to their ability to be a good symbiotic partner.
The Impact of Fermented Drinks on Antibiotic Resistant Genes in Adults’ Gut Microbiota
Name: Allen, Nyah
Email: nyaallen@iu.edu
Campus: IUB
Faculty Advisor: Dr. Funmilola Ayeni
Faculty Advisor's Email: fayeni@iu.edu
The emergence of antibiotic-resistant genes (ARGs) in human gut microbiota is a growing public
health concern. Antibiotics, essential for treating infections, can disrupt gut microbiota and
promote resistant bacterial strains. This issue is exacerbated by the slow development of new
antibiotics and the increasing prevalence of resistant infections. Fermented drinks like kefir and
yogurt, rich in probiotics, have been shown to positively influence gut microbiota. Probiotics can
restore gut balance, enhance gut barrier function, and inhibit pathogenic bacteria, potentially
reducing ARGs. This study aims to investigate whether regular consumption of fermented drinks
can reduce ARG prevalence in adults' gut microbiota.
A total of 36 participants (17 males and 19 females) from Nigeria were randomly assigned to
three groups: Control, Diet 1 (Millet drink - kunun zaki), and Diet 2 (Tigernut drink - kunun aya).
Fecal samples were collected at three stages: baseline, two weeks after the intervention, and
two weeks after the intervention stopped. Metagenomic DNA extraction from fecal samples was
performed using the QIAgen kit, followed by targeted PCR amplification for ARGs including
blaSHV, blaTEM, blaCTX-M, dfrA, qnrB, qnrS, qnrA, qepA, blaZ, ermA, ermB, mefA/E, and
aac(6’)/aph(2”).
Results indicated varying baseline prevalence of ARGs, with the blaTEM gene detected in 42%
of samples, conferring resistance to penicillin, and the dfrA gene in 78% of samples, reflecting
the widespread use of Septrin in Nigeria. No females had genes blaSHV, qnrB, qnrS, qepA,
aac(6’), ermB, and RPP tet detected. This study highlights the potential role of diet, specifically
fermented drinks, in modulating the gut microbiota and reducing the prevalence of ARGs.
Further analysis will focus on the comparative prevalence of ARGs between males and females
and the impact of fermented drinks on ARG presence in the gut microbiota.
Design of a Total Synthesis Pathway of the Precursors for the Molybdopterin Cofactor.
Name: Kibirige, Mark G
Email: mkibirig@iu.edu
Campus: IUI
Faculty Advisor: Dr. Partha Basu
Faculty Advisor's Email: basup@iu.edu
Molybdopterin is a cofactor for all essential forms of life. This cofactor is composed of a pterin, an ene-dithiolate, but does not have a molybdenum ion. A pterin is a bicyclic N-heterocycle that is found in many biomolecules. The precursors harbor 1,2- diketone or ozone functionality that have been condensed to 1,2 diamino benzene resulting in pterin derivatives following Gabriel Isay condensation reaction. Regioselectivity in this reaction was introduced by the addition of additives. The overall goal of this research is to learn about pterin synthesis to ultimately synthesize the molybdopterin cofactor. All compounds were characterized using 1H proton nuclear magnetic resonance and IR spectroscopies. This project will provide an opportunity to learn about these techniques.
Noon-12:15 p.m.: Break and pick-up boxed lunches & beverages
12:15-1:00 p.m.: Lunch & Panel Discussion: Nathan Alves, Paul Ardayfio, Rafael Bahamonde, Tabitha Hardy, Stephen Hundley, and Rebeca Mena
1:00-1:15 p.m.: Break
1:15-1:45 p.m.: Interactive discussions led by Colleen Manning
1:45-2:00 p.m.: Break and poster set-up (for session # 2)
2:00-2:45 p.m.: Poster Session #2
Upon arrival, poster presenter(s) will be directed to their poster board.
*Please note: Information about student poster presentations, including title and description, was provided by the individual student and/or their mentor(s).
Testing Protein Concentrations of Different Isomers: MDH1, MDH2, and PF Based on the Amount of IPTG in Solution
Name: Delgado, Elonzo
Email: epdelgado12@gmail.com
Campus: IUN
Faculty Advisor: Tia Walker
Faculty Advisor's Email: tialwalk@iu.edu
We are studying an enzyme where MDH is stable between a histidine tag. MDH has pathways in the body system within the Citric Acid Cycle so we are looking to manipulate MDH to bring some type of cure to pathogens.
Taming Bacillus: A Quest for the Perfect Culture Medium
Name: Dous, George
Email: gmdous@iu.edu
Campus: IUN
Faculty Advisor: Dr. Jen Fisher
Faculty Advisor's Email: fisherjc@iu.edu
In this study, we tackled the problem of Bacillus mycoides taking over culture plates when isolating bacteria from soil samples in Northwest Indiana. This overgrowth makes it difficult to find antibiotic-producing bacteria, which are crucial in the search for new antibiotics. To address this, we tested different types of culture media to see which ones can limit the growth of Bacillus mycoides while still allowing a variety of other bacteria to thrive. We experimented with several media, including 0.1x TSA, BHI Plates, R-2A agar, BUG Agar, BBL Simmons Citrate Agar, LB Broth, and Bacto Plate Count Agar. By finding the right medium, we hope to create the best conditions for discovering new antibiotic-producing microbes.
The Impacts of Hunting Enrichment on Larval Zebrafish (Danio rerio) Exploration and Stress Responses.
Name: Garcia, Ella
Email: eogarcia@iu.edu
Campus: IUN
Faculty Advisor: Maureen Rutherford
Faculty Advisor's Email: mlpetrun@iu.edu
Environmental enrichment has been shown to impact the behavior of zebrafish (Danio rerio) and increase stress resilience, but it is unclear whether hunting has the same effect as a form of enrichment. This study examines the impact of enrichment from live brine shrimp (Artemia nauplii) on the behavioral and physiological stress responses in larval zebrafish at 21 days post fertilization (dpf). In this work, larval zebrafish (5-21 dpf) were fed a live diet of brine shrimp or a commercial diet. At 21 dpf, the larva were subjected to a novel object test using a black Lego as the novel object. The larva were euthanized immediately after the novel object test to be used in future cortisol testing. It is expected that the sample fed live brine shrimp will show a decreased stress response to the novel object test and have lower cortisol levels when compared to the sample fed a commercial diet. These findings might suggest that live prey acts as enrichment for larva in a similar manner to environmental enrichment and therefore creates stress resilience in larva. Future studies should further examine the stress responses larval zebrafish undergo during hunting and the nutritional impacts of live food.
Lorentz Force Project
Name: Jennings, Jaylen
Email: jayjenn@iu.edu
Campus: IUN
Faculty Advisor:Jessica Warren
Faculty Advisor's Email: warrenjs@iu.edu
The overall goal of this project was to create an interactive VPython visualization of the Lorentz force law, a fundamental concept in electromagnetism. Understanding the Lorentz force is crucial for grasping the interactions between electric and magnetic fields, which are pivotal in numerous technological applications, from motors to particle accelerators.
Plasmodium Falciparum
Name: Ojwang, Natalya
Email: nojwang@iu.edu
Campus: IUN
Faculty Advisor: Dr. Tia Walker
Faculty Advisor's Email: tialwalk@iu.edu
In 2023, Malaria infections reacted 249 million cases with over 608,000 deaths. Resistance to antimalarial drugs is a challenge for effective treatment of Plasmodium falciparum (PF) infections. An individual is infected with Malaria when bitten by an infected Anopheles mosquito. PF is one of the deadliest parasites and is responsible for more than 90% of malaria deaths worldwide. New medications are needed in the fight against drug resistance. Malate dehydrogenase (MDH) is one of several enzymes found in the citric acid cycle, and protects against oxidative stress and in the transport of substrate through the metabolic pathways. PfMDH is an enzyme of interest for site selective inhibition, due to the ability to shut down active respiration. PFMDH was expressed in BL21DE3 E.coli using IPTG, extracted and purified through His-Tag/Ni-Agarose column chromatography, quantified by Bradford and analyzed by SDS-Page. Pure protein was subjected to specific activity assays including NADH/OAA affinity assays. Succinate is a metabolite in TCA and resent studies (March 2024 Bell Labs) suggest that Succinate may act as novel allosteric or orthosteric activator of MDH.
Positive Impact of C3 Grass Manipulation on the Growth of Forb Species in the Little Calumet River Prairie and Wetlands Preserve: Elymus virginicus L. and Four Forb Species
Name: Santiago, Alyna
Email: alysanti@iu.edu
Campus: IUN
Faculty Advisor: Dr. Spencer Cortwright
Faculty Advisor's Email: scortwr@iu.edu
Elymus virginicus L, commonly known as Virginia wildrye is grouped into a category well recognized as the C3 grasses. C3 refers to “cool season” grasses, which means that they thrive within climates that are cooler and range between 15-30℃. After the C3 grasses were given an ample amount of time to grow, a particular setup was crafted. Four different areas that contained the planted and grown Virginia wildrye grass within the Little Calumet River Prairie and Wetlands Preserve across from the Indiana University Northwest campus were used to conduct this study. Liatris pycnostachya (Lp), Baptisia lactea (Bl), Parthenium integrifolium (Wq), and Rudbeckia subtomentosa (Sbes) were the four forb species that were utilized within each of these four areas. The distance between each flowering plant was considered as well as the distance between each Virginia wildrye grass. Within two areas, cutting was done to specific C3 grasses to reduce the shading effect the dominant grasses had on the forb species. Our study indicated that the areas that contained the cut C3 grasses positively impacted the growth of the forb species. Rudbeckia subtomentosa was the forb species that was positively affected the most throughout this course of study.
Engineering a Fluorescent Biosensor for Real-time Monitoring of Ornithine in Bacterial Cultures
Name: Aljuboori, Zahra
Email: zaalju@iu.edu
Campus: IUSB
Faculty Advisor: Dr. Shahir Rizk
Faculty Advisor's Email: srizk@iu.edu
Ornithine, a non-canonical amino acid, is currently being studied for its role in bacterial interactions, specifically in the context of Enterococcus faecalis, which is known to secrete ornithine as a metabolic byproduct. This secretion has been shown to enhance the growth of P. aeruginosa. Using a biosensor, researchers could benefit by monitoring the presence of ornithine in real time. Ornithine binding protein (OBP), a periplasmic binding protein found in Salmonella, was used to engineer biosensors for ornithine. Four cysteine OBP mutants were constructed at strategic positions for attachment of thiol-reactive fluorescent tags, namely A22C, A54C, S120C, and A141C. OBP undergoes a conformational change upon binding ornithine, altering the fluorophore’s environment and thus induce a change in fluorescence. Our results show that the A22C mutant stands as the most strategically positioned mutant, as it’s the only mutant that demonstrated a discernible change in fluorescence in the presence of ornithine. Titrations of the various A22C mutant conjugates with ornithine resulted in dissociation constants ranging between 19µM and 2.9 mM. Furthermore, binding pocket mutations will be explored to increase selectivity for ornithine over arginine and lysine. The biosensors constructed here will be used to measure real-time ornithine concentrations in bacterial cultures.
Comparing Soil Physical/Chemical Properties and Biodiversity in Pinhook and Howard Park in South Bend, Indiana
Name: Barrera, Emily
Email: emsabarr@iu.edu
Campus: IUSB
Faculty Advisor: Dr. Deborah Marr
Faculty Advisor's Email: dmarr@iu.edu
High soil biodiversity in urban areas has been shown to benefit human health by reducing soil-borne pathogens and enhancing immune function. In addition, soils with low bacterial diversity tend to be dominated by methanogen bacteria that increase greenhouse gas emissions, and soils with low biodiversity (fewer species present) tend to have reduced ability to retain water and altered nutrient cycles. The purpose of this study was to collect baseline data on soil characteristics and soil invertebrate diversity in two South Bend city parks (Howard Park swales and Pinhook Park). In Fall 2023, for each park, we measured physical and nutrient properties of the soil including field capacity (ability of soil to hold water), percent soil organic matter, and nutrient levels including nitrogen, calcium, magnesium, and sulfate. We used a Berlese funnel to extract invertebrates from soil samples and calculated the diversity of invertebrates present at each site. Soil organic matter was more than two times higher at the Howard Park swales compared to the riparian zone along the shore of Pinhook lake. There were large differences in soil invertebrate diversity with Howard park having at least 17 invertebrates species compared to 4 invertebrate species documented in soil samples at Pinhook park. We also noted that plant species diversity was higher at Howard Park compared to Pinhook park. These data provided baseline information on soil health in city parks and can be used to inform landscape practices that improve soil ecological function and biodiversity.
Tracking Moth Diversity Trends: Analyzing Urban vs. Natural Habitats Across Two Years
Name: Bennett, Cordelia
Email: bennetco@iu.edu
Campus: IUSB
Faculty Advisor: Dr. Thomas Clark
Faculty Advisor's Email: tclark2@iu.edu
Moth diversity serves as a key indicator of habitat quality, given moths' vital roles in pollination and the conversion of plant biomass into animal biomass. This diversity faces growing threats from human activities, including habitat loss, reduced availability of native plants for caterpillars, and light pollution, which collectively has led to significant declines in moth populations. Building on a summer 2023 study that examined variations in moth species abundance across urban, urban-restored, and natural sites, this study investigates changes over the course of one year at two sites: Potato Creek (natural) and Indiana University Wetlands (urban). The 2023 study revealed moth larvae who were internal feeders (mostly microlepidoptera) were more prevalent in urban areas, while those more exposed with external feeding (mostly macrolepidoptera) thrived in natural settings. Using our current research findings and last year’s results we will assess the percentage of species that persisted, emerged, and any shifts in the macro/micro species ratio. This analysis aims to provide insights into strategies for reversing declines in urban moth diversity, validate or challenge previous findings, and provide a comprehensive moth census for the studied locations in order contribute to the natural history knowledge of the chosen preservers.
Environmental Conservation of Moths
Name: Cardenas, Guillermo
Email: Gucarden@iu.edu
Campus: IUSB
Faculty Advisor: Dr. Thomas Clark
Faculty Advisor's Email: gmuna@iu.edu
We wanted to study the moths' survivability in urban environments and compare them to data that we got last summer and see what species are thriving and which are not. In order to answer our question we had to find a way to trap the moths without harming them. For this we came up with a light trap that attracts the moths to a white sheet we set up, as they landed it gave us the opportunity to take a photograph of the moths for us to later identify off site. A total of 5 different sites were used for the study, two at Potato Creek, two at a Lydick Bog, and one at a wetlands. With our data we were able to see that the moths that laid their larvae in hidden or concealed spaces do better in urban environments as opposed to those that have exposed larvae.
A Course-Based Undergraduate Research Project: A Correlation Study of Lead Levels in Contaminated Soil Samples with Source, Home Location, and Age.
Name: Conn, Megan
Email: connm@iu.edu
Campus: IUSB
Faculty Advisor: Dr. Grace Muna
Faculty Advisor's Email: gmuna@iu.edu
Lead is a toxic heavy metal that is present in the environment due to human activities. The major source of lead contamination is from the use of leaded gasoline and lead-based paint for many decades before it was banned. Although lead is no longer used, once deposited it moves very little through the soil and can persist for a long time. The research goal was to correlate lead levels in soils in several homes with the location, source of the soil whether from the front yard or back yard, and the year the homes were built. Lead was extracted using a modified EPA method 3050B and analyzed using microwave plasma atomic emission spectroscopy at 405.78 nm. The data show a correlation between the amount of the lead in the soil with home location and age. Homes located in the city show higher amount of lead compared to homes located in the farms or in newly developed areas. In addition, there is also a good correlation between the levels of lead in soil samples collected from the front yard and those obtained from the back yard in homes located in the city, front yard showing slightly higher levels.
Developing a Sensitive Electrochemical Sensor for Detecting Cortisol using a Gold Electrode Modified with Gold Nanostructures
Name: Cortez Flores, Edwin
Email: ecortezf@iu.edu
Campus: IUSB
Faculty Advisor: Dr. Grace Muna
Faculty Advisor's Email: gmuna@iu.edu
Constant exposure to stress leads to the production of cortisol, a major risk factor and a silent contributor to various health-related disorders. Therefore, monitoring cortisol levels is crucial in understanding its biological and physiological effects, including blood glucose levels, blood pressure, immune system responses, and other health conditions such as Cushing Syndrome or Addison’s disease. This work involves modifying a gold electrode with gold nanostructures deposited electrochemically. A self-assembled monolayer of dithiobis (succinimidyl) propionate (DTSP) was prepared on the gold nanostructures, by deposition on the gold nanostructures. DTSP forms a strong Au-S bond with the gold through the cleaved sulfide bonds. This creates a surface that can be used to immobilize the cortisol antibodies through the primary amine group and the amine-reactive N-hydroxysuccinimide ester on the DTSP molecule creating a surface to detect cortisol through binding. The detection of cortisol was monitored using cyclic voltammetry and differential pulse voltammetry techniques. Future work will focus on determining the limit of detection (LOD), stability, and selectivity of the developed method as well as testing its performance in determining cortisol in real samples.
Comparison of Soil Ecosystem Function Across South Bend Urban Tree Nursery Sites
Name: Diaz, Perla
Email: Prdiaz@iu.edu
Campus: IUSB
Faculty Advisor: Deb Marr
Faculty Advisor's Email: dmarr@iu.edu
The city of South Bend is working to increase urban tree canopy (UTC) coverage to 40%, focusing on underserved neighborhoods. We measured soil biodiversity and soil properties in proposed tree nursery sites and campus sites to establish baseline data. To compare soil quality, we measured soil invertebrate diversity, soil pH, heavy metals (Pb, Cu, Cd, Zn, Mn), carbon content, water-holding capacity, and decomposition rates at each site. Our findings showed that the campus wetland with more native plants and had higher soil invertebrate diversity compared to sites with more nonnative vegetation. Metal levels were variable across sites. However, campus sites had lower lead and lower pH compared to city sites. Sites with higher lead concentrations had 20-30% abundance of soil mites, and sites with low lead concentrations had 10-15% mites. Studies have shown that some species of soil mites can tolerate lead. Sites with higher water capacity tended to have higher percent carbon. We observed higher decomposition in green tea bags compared to rooibos indicating more bacterial activity compared to fungal. These data are relevant to the city's urban tree project because high soil biodiversity, water-holding capacity, and other soil properties affect tree survival and growth.
Modulating PhuZ Polymerization Dynamics with Periplasmic Binding Proteins
Name: Hayes, Elizabeth
Email: elizhaye@iu.edu
Campus: IUSB
Faculty Advisor: Dr. Shahir Rizk
Faculty Advisor's Email: srizk@iu.edu
Elizabeth Hayes, Zahra Aljuboori, and Shahir S. Rizk
Department of Chemistry and Biochemistry, Indiana University South Bend
Microtubule systems are key cytoskeletal components in eukaryotic organisms. Studies support the presence of similar systems in large bacteriophage. PhuZ, a tubulin-like protein, organizes into long polymers when bound with guanosine triphosphate (GTP) due to a change from an open to closed structural conformation. The ligand triggering the polymerization activity of PhuZ may be modified when linking periplasmic binding proteins (PBP) to the N-terminus. Similarly, PBPs exist in various conformations depending on their ligand-binding state. In this study, fusions of maltose binding protein (MBP) and glucose binding protein (GBP) were assessed using AlphaFold, an AI molecular modeling tool. UCSF Chimera provided visualization of these models. Current goals are to construct these fusions using PCR mutagenesis. Then, the constructs are to be validated with DNA sequencing and expressed in E. coli. If successful, there will be a new visual detection system measured by the concentration of each PBP’s respective signal. Additionally, PhuZ may offer insight into microtubule dynamics and allow for re-engineering of the forces that trigger molecular assembly contributing to the development of novel bio-responsive biomaterials.
Electrochemical Detection of 3-Nitrotyrosine using a Gold Electrode Modified with N-hydroxysuccinimide Ester Gold Nanoparticles
Name: Johnson, Luke Johnson
Email: LukeJohnson1506@gmail.com
Campus: IUSB
Faculty Advisor: Grace Muna
Faculty Advisor's Email: gmuna@iu.edu
3-Nitrotyrosine (3-NT) results from the nitration mediated by reactive nitrogen species such as peroxynitrite anion and nitrogen dioxide. It is overproduced when there is oxidative stress in the body, which is an imbalance of free radicals and antioxidants within the body. Oxidative stress is common in many diseases like cancer and heart disease. 3-NT has been identified as a biomarker for cell damage, inflammation, as well as nitric oxide production. Therefore, its monitoring is important for disease monitoring. An electrochemical biosensor platform was prepared by modifying a gold electrode with N-hydroxysuccinimide (NHS) ester gold nanoparticles. The NHS ester gold nanoparticles created a monolayer on the electrode surface which was used to covalently immobilize 3-NT antibodies. The modified electrode surface was then used for the detection of 3-NT using cyclic voltammetry and differential pulse voltammetry. The detection method was very sensitive to 3-Nitrotyrosine with a linear response from 50 to 600 nM concentrations. Future research will involve testing the selectivity, and stability of the method as well as testing real biological samples such as urine, serum, and saliva for potential practical applications.
The Brilliance of Periplasmic Binding Proteins: Illuminating the Future of Nutrient Monitoring in Bacterial Culture
Name: Malkovsky, Joseph
Email: jmalkovs@iu.edu
Campus: IUSB
Faculty Advisor: Dr. Shahir Rizk
Faculty Advisor's Email: biochemnerd@gmail.com / srizk@iu.edu
The goal of our research was to monitor nutrient consumption by bacteria in culture in real-time. We used mutant periplasmic binding protein (PBP), which are used by bacteria to obtain resources in a competitive environment. Arabinose binding protein (ABP) captures arabinose and undergoes a conformational change from an open to a closed form. Previous studies have tested attaching various fluorophores at different locations on PBPs to evaluate the greatest change in fluorescence between the open and bound conformation. We adopted those locations and attached some of the same fluorophores but included Alexa Fluor 488 (AF488). We attached the fluorophore by substituting a cysteine into the sequence of ABP at position 253 of the amino acid sequence, to form a covalent bond between ABP and each fluorophore. We monitored the change in fluorescence of the ABP-fluorophore conjugates at increasing concentrations of arabinose to find an ideal candidate to act as a fluorescent biosensor for the sugar. ABP253C-AF488 exhibited the greatest fluorescence change from 524nm, with a KD of 28.2 ± 1.45 μM. We tested ABP253C-AF488 in growing cultures of Escherichia coli and monitored arabinose consumption. This method could be used to generate different proteins to monitor multiple nutrients in bacterial culture concurrently.
Assessment of Metal Contamination in Soil: A Correlation Study of Metal Ion Levels with Source, Home Location and Age.
Name: Mukeshkumar Chaudhari, Dhruval
Email: dhchaudh@iu.edu
Campus: IUSB
Faculty Advisor: Dr. Grace Muna
Faculty Advisor's Email: gmuna@iu.edu
The presence of metal ions such as lead, copper, zinc and manganese in high concentrations in the soil can be a source of pollution. For example, lead is a potent neurotoxin with no known safe level of exposure, making it a significant public health concern. High concentrations of zinc in the human body interferes with the absorption of other essential minerals and disrupts cellular processes. Copper is toxic when excess in the body because it disrupts cellular processes by causing oxidative stress and DNA damage. Elevated levels of manganese can potentially cause neurological disorders that affect brain development in children. The goal of the present work was to extract and analyze metal ions in soils from several homes within the Michiana area to gauge the level of metal contamination. The samples were extracted using a modified EPA method 3050B and analyzed using microwave plasma atomic emission spectroscopy. The results show a correlation between metal ion levels with the location, source, and the age of the homes. Homes located in the city show higher amounts of metal ions compared to homes located on farms or in newly developed areas. Older homes had higher levels of metal ions than newer homes.
Reactions and Electrochemistry of Naphthoquinones and Sulfide
Name: Nyirongo, Moses
Email: mnyirong@iu.edu
Campus: IUSB
Faculty Advisor: Kasey Clear
Faculty Advisor's Email: kclear@iu.edu
1,4-Napthoquinone and its derivatives, Juglone, Menadione, DMNQ, and HMNQ, collectively known as NQs, are naturally occurring and synthetic species that can have therapeutic applications when used at lower levels. They are attributed to redox cycling and the production of reactive oxygen species. When reacted with H2S, it is known that NQs are reduced into semiquinones and hydroquinones, via a one or two-electron transfer, respectively. It is unknown if NQs form an adduct during. For this research, we explored NQs and their oxidative properties on hydrogen sulfide (H2S) into reactive sulfur species. We reacted NQs with Na2S in aerobic conditions and performed thin layer chromatography (TLC) analysis which showed that the reaction resulted in the formation of new NQ species. We also performed cyclic voltammetry (CV), and the results showed that the reaction was diffusion-controlled as the reduction peak current decreased as we increased the voltage of the scan rates over time. With the CV results, it can be observed that an adduct that is no longer accessible for electron transfer forms.
Quantification of Polyphosphate in Bifidobacterium using Fluorescence and Non-fluorescence Dyes
Name: Robinson, Benjamin
Email: benrobi@iu.edu
Campus: IUSB
Faculty Advisor: Yilei Qian
Faculty Advisor's Email: yilqian@iu.edu
Abstract:
Bifidobacterium is a genus of gram-positive bacteria that are known to be the beneficial colonizers of the human intestines. Being strictly anaerobic, they are typically limited to the gastrointestinal tract (GIT) region, but some are also found outside the GIT. Bifidobacterium dentium, for example, was found in the oral cavity and isolated from dental carries. Previously, it was observed that, compared to other bifidobacterial species that are exclusive dwellers of the GIT, B. dentium cells exhibited a higher basal level of polyphosphate (PolyP) granule production, which is associated with acid-tolerance in this organism and other bacteria. We hypothesized that a high level of polyphosphate granules plays an important role in B. dentium’s survival in extra-intestinal spaces, such as dental caries. Therefore, quantification of PolyP production is important to studying its role in its environmental adaptation and survival. In this study, we developed a fluorescence method to quantify PolyP production in B. dentium and another GIT species, Bifidobacterium longum, using 4′,6-diamidino-2-phenylindole (DAPI), a fluorescent dye that can bind to DNA and PolyP. Our results showed that B. dentium produced a similar high level of PolyP in both PolyP-inducing medium (AMS medium) and the control medium. In B. longum, however, PolyP production was only produced in the AMS medium. We also developed a differential staining method using chromophore dyes which showed a dose-dependent response in PolyP production. To conclude, the fluorescence and non-fluorescence method can both be used to quantify PolyP production in bifidobacteria.
Acid Tolerance and Resistance to Oxidative Stress in Oral Bifidobacterium and Streptococcus Species
Name: Rojas Romero, Ashley
Email: arojasro@iu.edu
Campus: IUSB
Faculty Advisor: Yilei Qian
Faculty Advisor's Email: yilqian@iu.edu
Bifidobacterium is a genus of beneficial bacteria that primarily reside in the human gastrointestinal tract, but some species have also been found in the oral cavity. One such species, Bifidobacterium dentium, has been found not only in the intestines but in dental caries as well. Compared to other narrow-range intestinal bifidobacterial species, B. dentium produced a high level of polyphosphate granule (PolyP) in sucrose, even under reducing conditions. We hypothesize that its PolyP-producing ability could contribute to its resistance to oxidative stress and acidic pH in the oral cavity. This hypothesis was tested in three species: B. dentium, Bifidobacterium longum (intestine-only), and Streptococcus mutans (a common cariogenic pathogen). Culture media were designed to mimic oral cariogenic conditions, such as the use of sucrose (“table sugar”), and calcium phosphate (the major component of tooth enamel). Acid-tolerance and oxidative stress experiments were carried out in these three organisms. Results indicated B. dentium had a higher acid tolerance than B. longum and S. mutans under both PolyP-producing and non-producing conditions. The granule-producing cells of B. dentium and B. longum were also more acid-resistant than the non-granule-producing cells. The two bifidobacterial species also demonstrated a greater resistance to hydrogen peroxide under granule-producing conditions. S. mutans, an aerotolerant species, was the most resistant to hydrogen peroxide. We concluded that the high-level granule production in B. dentium might contribute to its survival in unfavorable environments outside the intestines.
Engineering a Biosensor for Lactate, a Marker for Inflammation and Bacterial Fermentation.
Name: Sonnenberg, Alissa
Email: alsonnen@iu.edu
Campus: IUSB
Faculty Advisor: Dr. Shahir Rizk
Faculty Advisor's Email: srizk@iu.edu
Lactate levels within the human body can be indicative of inflammation or injury. Lactate is also a byproduct of fermentation and therefore, its levels can be used to assess if foods are properly preserved. Here, we engineer a protein-based biosensor for lactate to improve how lactate is measured and monitored. To accomplish this, we utilized a bacterial lactate binding protein from Thermus thermophilus. We constructed mutations to introduce individual cysteines at specific positions for attachment of thiol-reactive fluorophores. After identifying the cysteine mutation and fluorophore combination that showed the greatest fluorescence change in response to lactate binding, we carried out titrations with lactate to determine the dissociation constant (Kd) of the protein for lactate. The Kd, which indicates the midpoint of the concentration range the biosensor is able to detect lactate, was 1.4 µM, reflecting the high affinity of the biosensor for lactate. These results, and results from experiments utilizing artificial sweat, fetal bovine serum, and pickle and olive juice, suggest that our biosensor could identify stress in the body, and also test foods to ensure they’ve been adequately preserved. Further research should be conducted to test the biosensor’s abilities in human sweat and blood.
Constructing Fluorescent Biosensors to Understand Bacterial Metabolites
Name: Underdue, Morgan
Email: munderdu@iu.edu
Campus: IUSB
Faculty Advisor: Dr. Shahir Rizk
Faculty Advisor's Email: srizk@iu.edu
One way to understand pathogenic bacteria is through studying how they consume different metabolites under different conditions. Our goal is to use periplasmic binding proteins (PBPs) conjugated to fluorophores as biosensors to monitor specific bacterial metabolites in real-time. PBPs undergo large conformational changes when bound to their ligands. By mutating these proteins and attaching environmentally sensitive fluorophores in specific locations, fluorescence can be used to monitor the presence of the PBP’s ligand. Here, we use glutamate binding protein (EBP) mutants (EBP126C and EBP207C) conjugated with fluorophores (acrylodan, Alexa-488, coumarin, or IANBD) to detect the levels of glutamate in culture. To determine the optimal range of glutamate detection, the dissociation constant for EBP126C-acrylodan was determined to be 0.35µM and 6.42mM for EBP126C-coumarin. With a 43% decrease in fluorescence upon glutamate binding, our results show that the EBP126C-coumarin conjugate is ideal for detecting changes in glutamate concentration in bacterial cultures. Combined with other periplasmic binding proteins, our approach can be used to detect changes in multiple analytes throughout the growth of bacteria in real-time. Monitoring these changes will be helpful in studying the intricate relationships between bacterial metabolite use and pathogenesis.
2:45-3:00 p.m.: Wrap-up and concluding remarks
3:00 p.m.: Adjournment

The Louis Stokes STEM Pathways Implementation-Only Alliance: Indiana LSAMP is funded by the National Science Foundation, award number EES 2308500, 2023-2028. Any opinions, findings, and conclusions or recommendations expressed in the contents of this website are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.